Genotype Download using Search Wizard
====
## Related Genotype Data
The Search Wizard provides an easy way to download genotype data. You are required to login to use the Search Wizard because the tool uses saved lists and datasets that are linked to an account. After selecting the data, you find the download link in "Related Genotype Data". The features it provides are:
1. Selection by Genotype Protocols, Genotype Projects, Breeding Program, Year, Location, Accessions, or Seedlots.
2. You can also start your selection with a list of accessions or a predefined dataset.
3. The output format of genotype data is VCF or Dosage matrix.
4. The output can be filtered by chromosome, position, or markerset.
### Performance
The limitations of using the Search Wizard are that it becomes too slow when downloading over 1M genotypes. So, if you are downloading 90K data you are limited to 50 accessions or if you are downloading a large exome capture protocol you may only be able to download a few accessions at a time. For larger downloads use the "Archived Genotype Data" method for download.
### Requirements
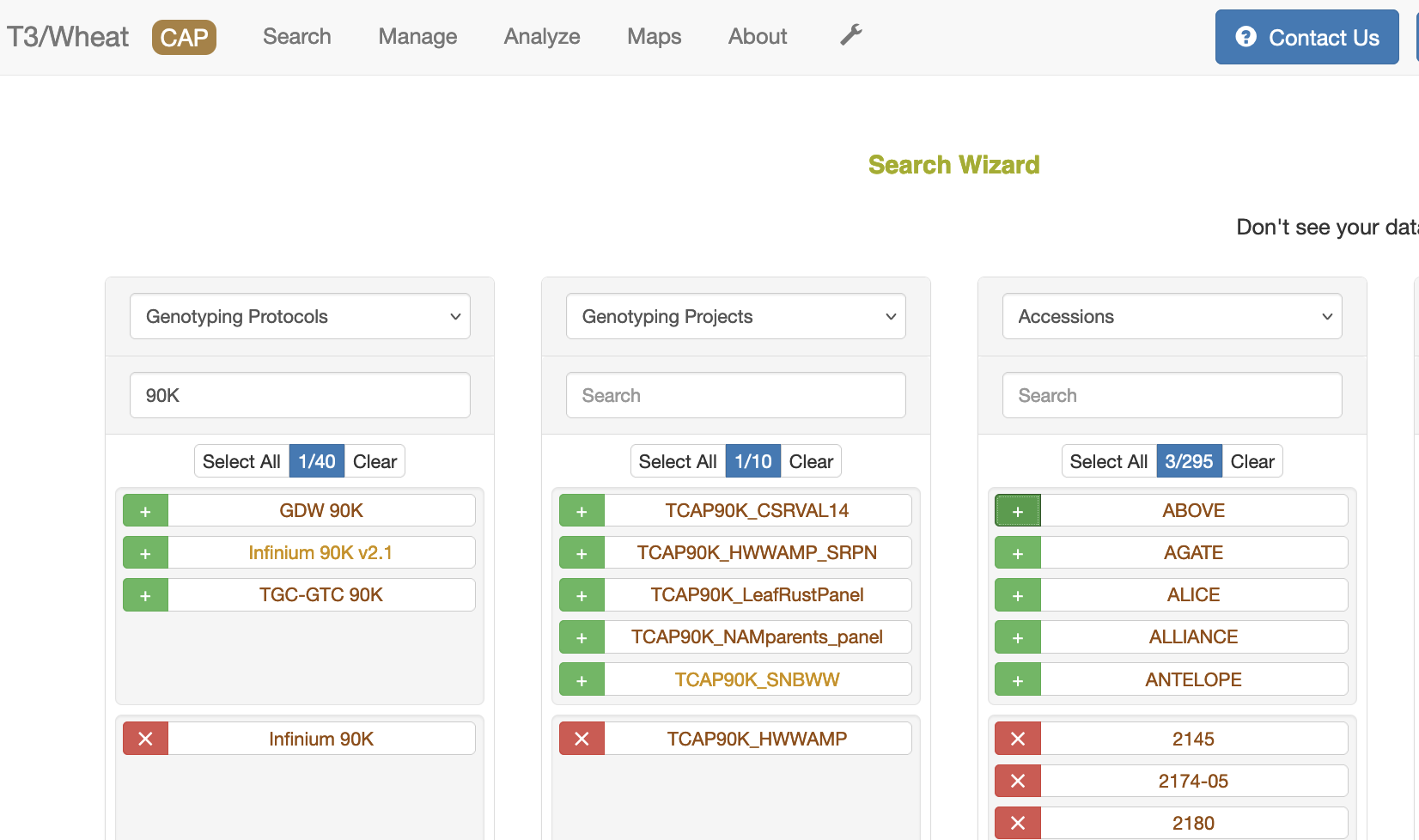
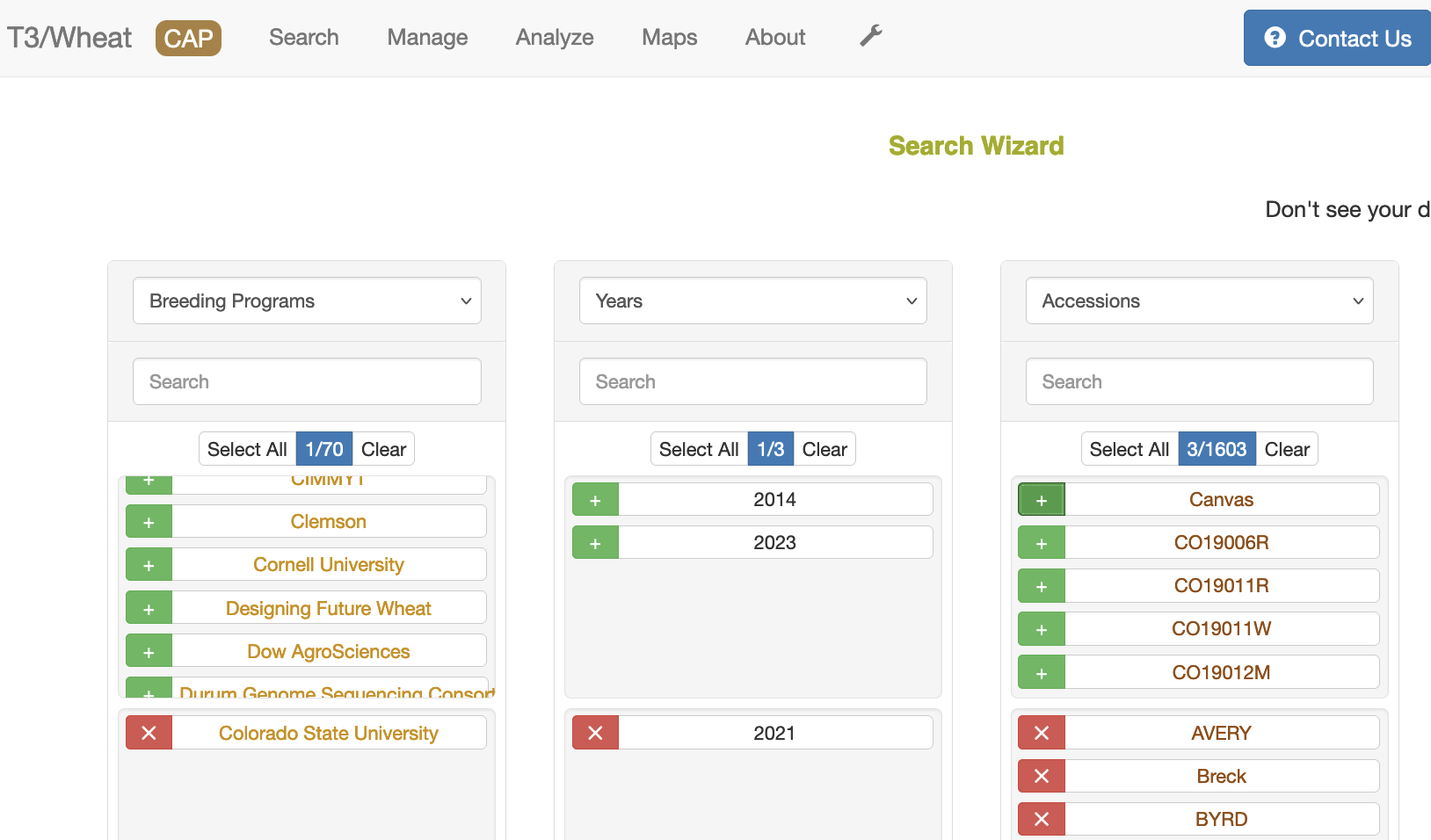
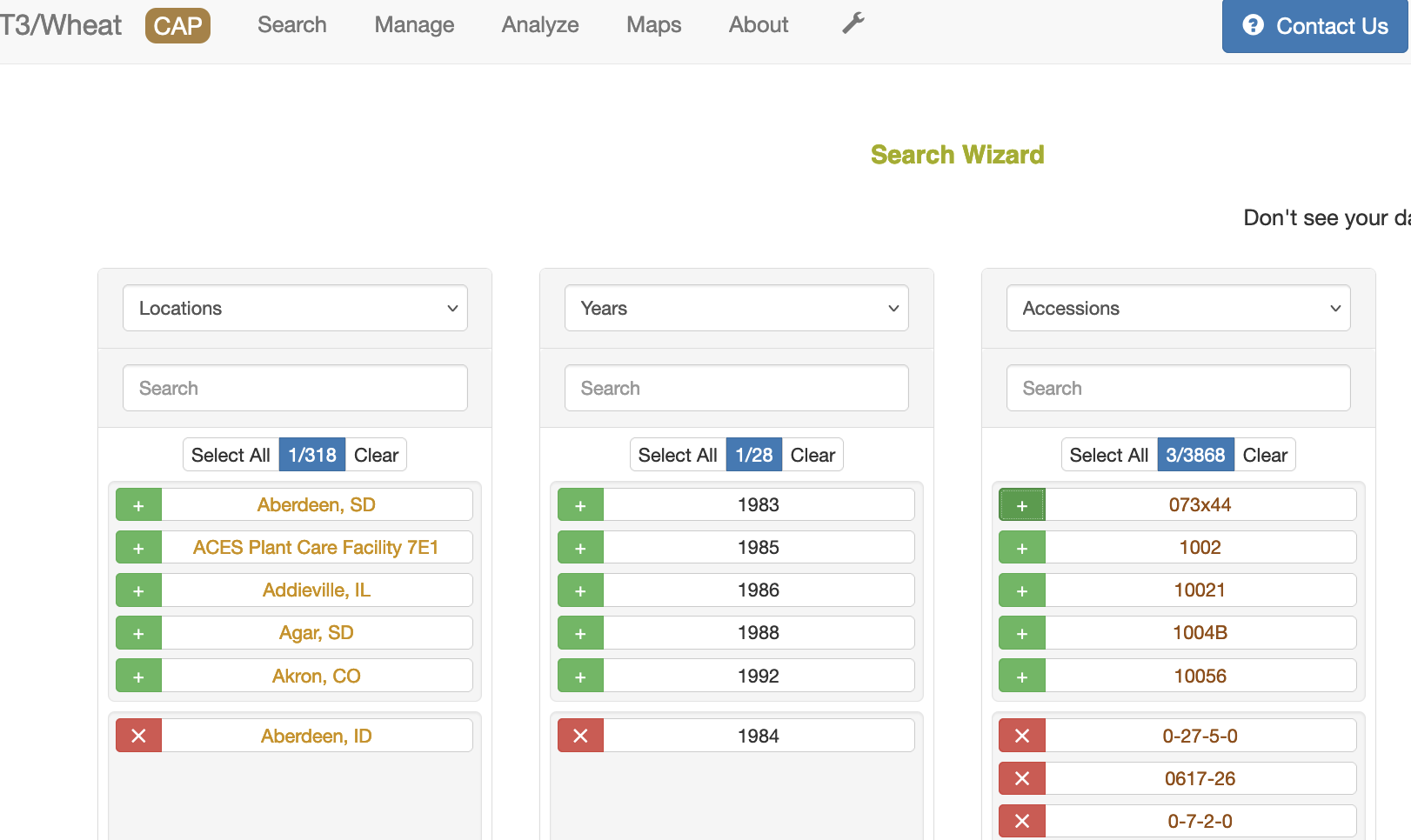
The only requirement for downloading genotype data is to have accessions selected. First, select the data type in the column heading, then select the green "+"" sign to select a specific entry. If you select the name of the entry, you will open another tab with details on that entry. You can type the name of the entry you are looking for to filter the list of entries. You can usually select more than one entry except for genotype protocol. Selecting more than one genotype protocol will disable the download option.
### Protocol Assumptions
If no genotype protocol or project is selected, then the wizard uses a default protocol (90K) which is listed in the bottom of the Related Genotype Data section. If a genotype project is selected, then the genotype protocol associated with that project is used. If a genotype protocol is selected, then the download only contains data from that protocol.
### Instructions
The general procedure for using the wizard is to start with the most general category in the left column then refine the choice in the following columns. Some examples of this are:
1. Genotype protocol in the first column, then genotype project, then accessions.
2. Breeding program in first column, year in second column, then accessions
3. Location in first column, year in second column, then accessions
After selecting, click on the “Related Genotype Data” button, then click on "Download Genotypes".
**Example Search Wizard Selections:**
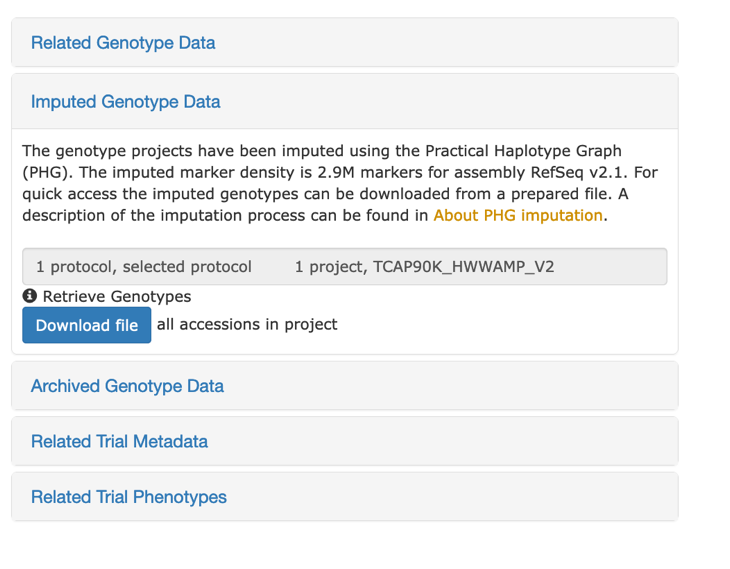
## Imputed Genotype Data
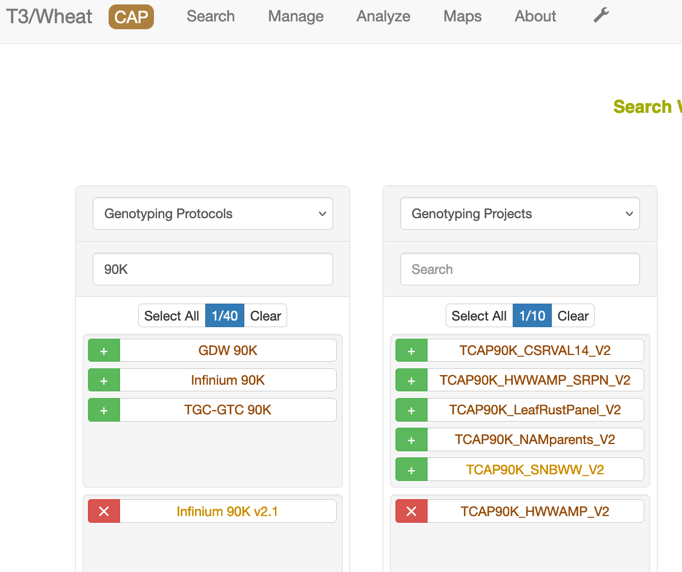
For the T3/Wheat website some genotype projects have been imputed. This genotype data download requires that you have a genotype protocol and genotype project selected. Each genotype project has been imputed using the Practical Haplotype Graph software v0.4 using a database of 472 wheat cultivars and the RefSeq v2.1 assembly. The imputed data contains 2.89M markers. The data is provided in a gzipped VCF file. Select a genotype protocol (with RefSeq v2.1) and a genotype project, then click on "Imputed Genotype Data" button then click on "Download File".
## Archived Genotype Data
This download option was created to quickly download large datasets. You will get a VCF file that was loaded into T3. This genotype data download requires that you have genotype protocol or genotype project selected.
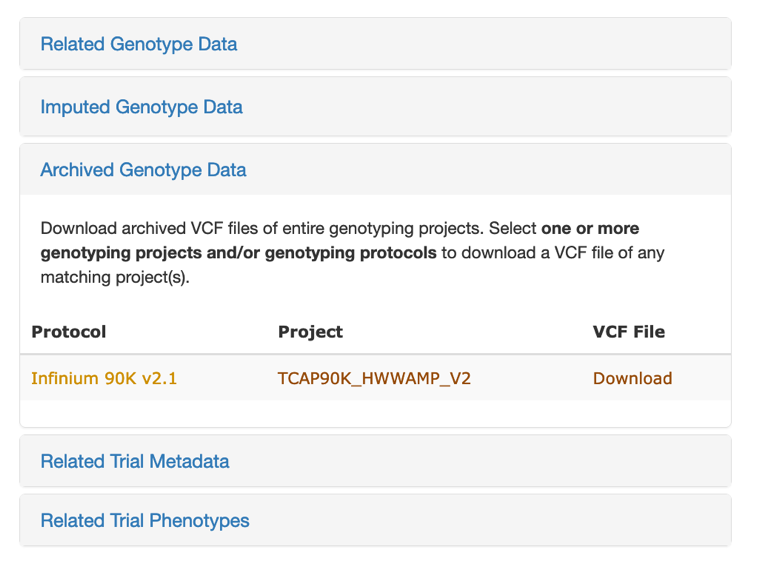
Archived VCF files can also be downloaded from the Genotyping Project detail page (accessible from the Search > Genotyping Projects page).
## Additional Options
### Filtering by Chromosome and Position
The "Related Genotype Data" section provides an option for filtering the downloaded data. You can select a specific chromosome and limit the data to a specific location on that chromosome.
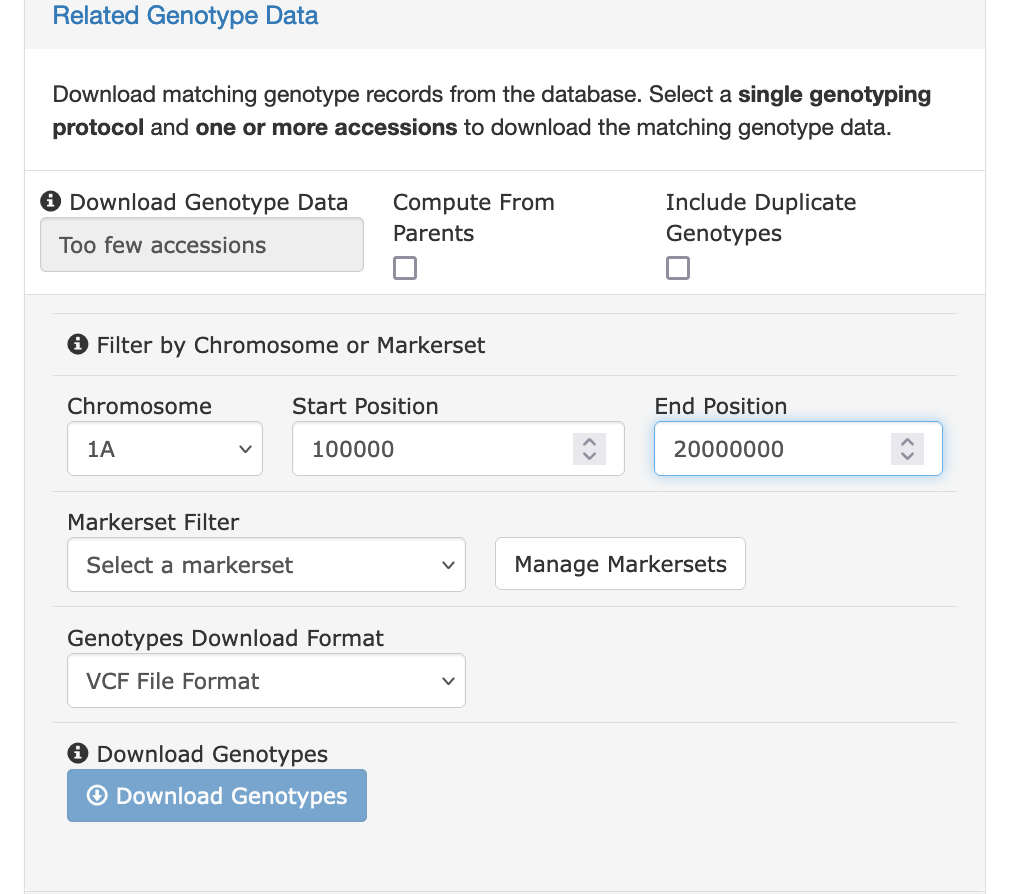
### Filtering by Markerset
The "Related Genotype Data" section provides an option for filtering the downloaded data for specific markers. Before using this feature, you first must
1. Navigate to Manage => Markerset
2. Add new markerset
3. Add markers to a markerset
Then return to the wizard and select the created markerset in the "Related Genotype Data" section.
### Using Lists
For the first column in the search wizard you can select a saved list to populate the selection or create a new list from the selection. In the area where you select the column type, if you scroll down, you can see a section titled "Load Selection from List". The lists are created either within the wizard or using the manage lists button on the upper right of the web page.
### Using Datasets
A dataset is a collection of lists of dieerent types. For example, if you select a protocol, project, and accessions you can save all those selections into a dataset for retrieval in the wizard or for use in many of the analysis tools. The datasets are loaded in the "Load/Create Datasets" section of the wizard page. You can view the details of all the datasets using the manage datasets button on the upper right of the web page.